NANOMICS
NAnopore sequeNcing for Operational Marine genomICS

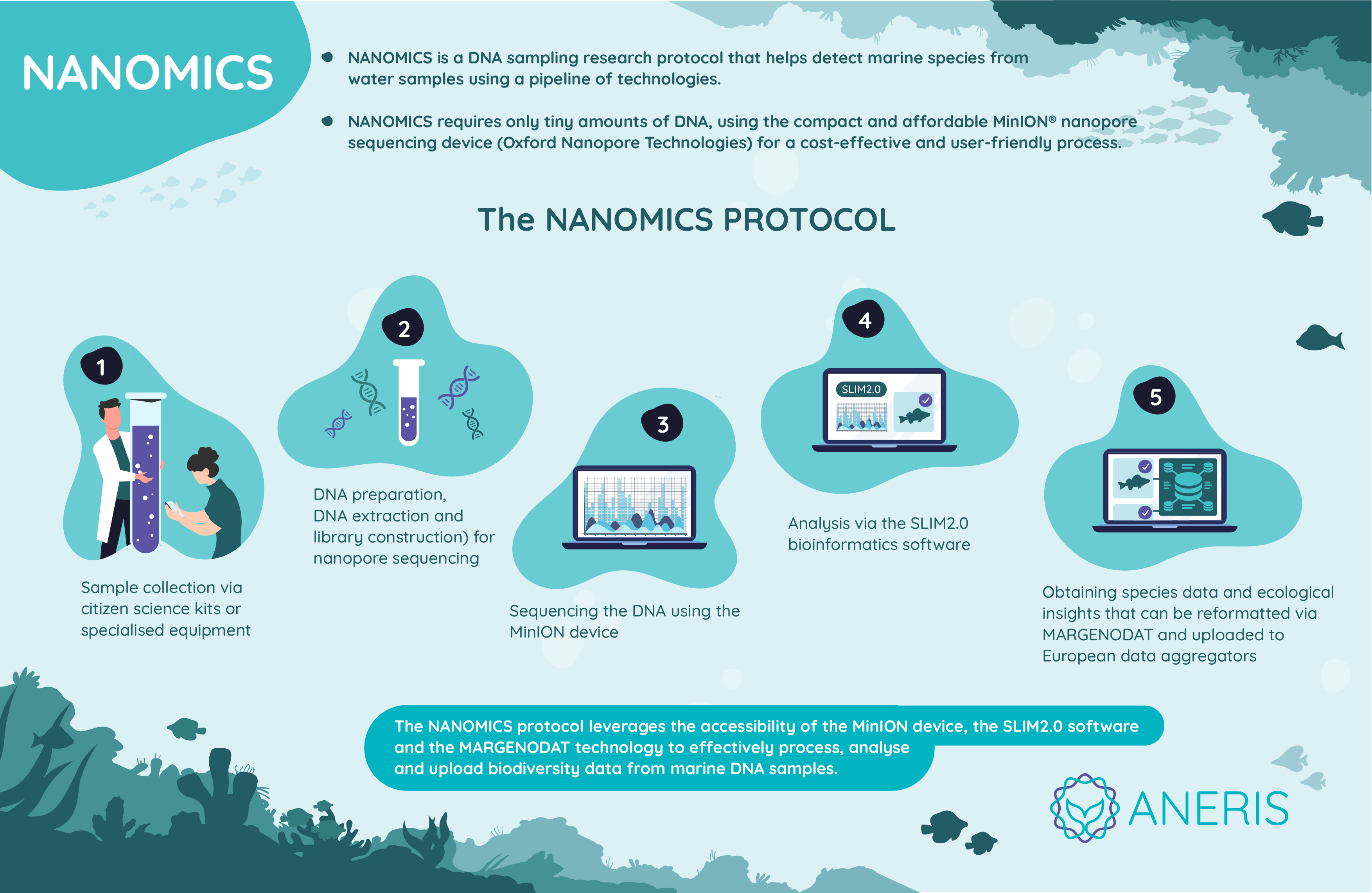
The current state of high-throughput genetic biodiversity assessment (i.e DNA metabarcoding) revolves mostly around Illumina Next Generation Sequencing (NGS) technology, which cannot be applied in field conditions.
Applying a Nanopore sequencing technology on a portable device can be much more effective in terms of capabilities, cost, delivery of results and applicability in the field.
ANERIS plans to develop, test and optimise Nanopore sequencing protocols for different taxonomic groups (e.g. fish, plankton or rocky benthic environmental DNA) and compare them to standard Illumina metabarcoding data.
Currently, NANOMICS is focused on creating and evaluating sampling protocols for the different study sites and ecosystems (water samples, intertidal, sediments, etc.
Target Stakeholders
Governmental Authorities: Water Framework directive, Ocean Best practices from UNESCO
Citizens, NGOs, associations: Natuurpunt, engaged citizens, WILDERLAB, UNESCO eDNA
Academia: Rbins, OBON: Ocean Biomolecular Observation Network (UN Ocean Decade action), EMBRC stations are interested in protocol development (e.g. VLIZ and HCMR in ANERIS)
Industries: Illumina, Oxford Nanopore Technologies



